Using genetics to build a better understanding of dementia
The causes of dementia are usually complex and involve a combination of genetic, environmental and lifestyle factors. Thanks to advances in DNA sequencing technologies, we have now discovered several genetic variations that can influence a person’s chance of developing a disease. Identifying these so-called ‘risk genes’ is providing incredible insight into the biological mechanisms involved in tipping the balance from health to disease.
In fact, with many risk genes for neurodegenerative diseases already uncovered, scientists are beginning to investigate the key relationships between them in order to improve our understanding of disease mechanisms. In addition, these studies help us gain a holistic view of how the genes interact so that we ascertain the chance that an individual will develop one of these conditions.
The Hardy Lab is aiming to build complex disease networks for different neurodegenerative diseases. Their work involves using bioinformatic approaches to analyse gene activity and genetic data from cells, human brain tissue and laboratory models. The Hardy Lab are working closely with other researchers to test the predicted networks through laboratory experiments in cells and tissues. They hope this approach will ultimately lead to the development of new treatments that can stop, slow down or reverse dementia.
Latest news


Prof Sir John Hardy
Prof Sir John Hardy is a Group Leader at the UK DRI at UCL. Find out more about his career and expertise on his profile page.

Research summary
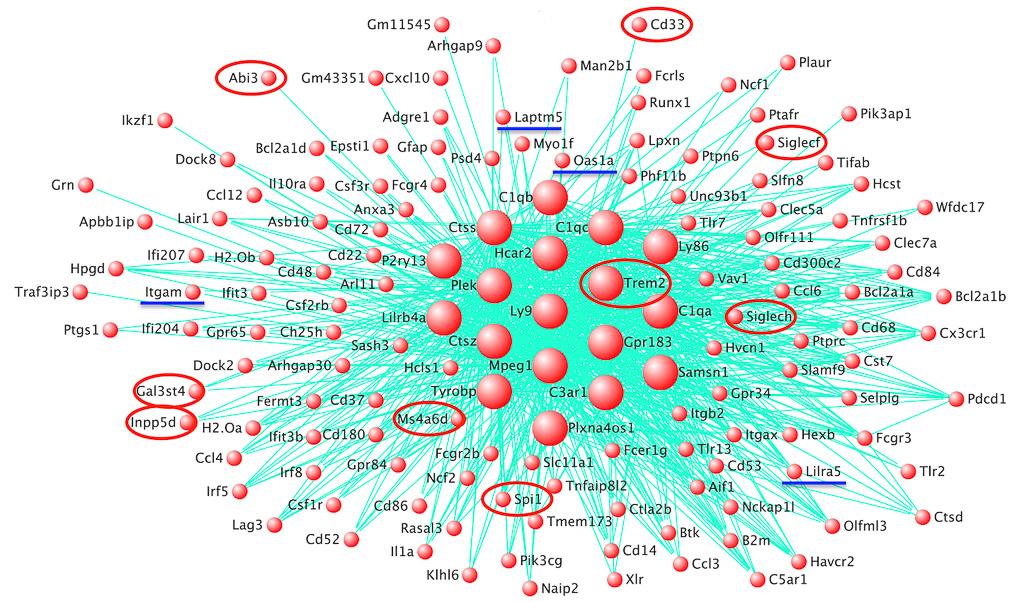
The Hardy & Escott-Price Labs identified four new risk genes for Alzheimer’s disease, tied to the microglial response to amyloid beta. Salih et al, 2019
Building pathogenic networks for neurodegenerative diseases
In the major neurodegenerative diseases, discoveries in genetics, and the following analysis has traditionally been valued solely as a driver for molecular investigation. However, with the massive increase in identified genetic loci driven by GWAS and by whole-exome and whole-genome sequencing, the balance of information transfer is shifting: so many genetic loci have now been discovered for the major neurodegenerative diseases (Alzheimer’s disease, Parkinson’s disease, Motor Neuron Disease and to a lesser extent frontotemporal dementia (FTD)) that we are now able to distinguish emerging networks of genes involved in pathogenesis. Importantly, these networks now allow us to have a better understanding of genetic risk, not just in terms of individual genes but in genetic variability in processes driving pathogenesis.
It is now possible to use bioinformatics to predict which particular genes and pathways are likely to be involved in neurodegenerative diseases – the expertise of the Hardy Lab. The first successful example of this was the identification of the TREM2 network in amyloid response in mice, pointing to the new genetic loci PLCG2 and ABI3. Results from the team have also shown that nearly all the pure FTD genes are lysosomal, nearly all the FTD/ALS genes belong to the ubiquitin-proteasome pathway and that ataxia genes are likely to be involved in calcium homeostasis, the ubiquitin-proteasome system or DNA repair.
This programme will involve the use of whole-genome expression analysis in cells, mouse models and human brains. The bioinformatic analysis that follows will help guide ongoing genetic investigation in these diseases, in order to unravel disease pathways. This bioinformatic information will be shared with collaborators working on cell and animal systems, and those working on disease biomarkers, to drive a programme of work designed to advance our understanding of disease pathogenesis and devise novel therapeutic approaches.
Main objective and research goal:
The overall aim is to build pathogenic networks for neurodegenerative diseases based on network analyses of expression and genetic data and then to begin to test these networks through experimental work in relevant tissues and cells in collaboration with others.
Key publications
Vacancies
Lab members
- June Smalley (PA)
- Dr Anupriya Dalmia (Senior Researcher)
- Dr Kin Mok (Postdoctoral Researcher)
- Dr Dervis Salih (Postdoctoral Researcher)
- Dr Maryam Shoai (Postdoctoral Researcher)
- Dr Melissa Barber (Postdoctoral Researcher)
- Anna Staub (Postdoctoral Researcher)
- Amanda Assoni (Postdoctoral Researcher)
- Umran Yaman (PhD Student)
- Naciye Magusali (PhD Student)
- Hannah MacPherson (PhD Student - Jointly with Mina Ryten)
- Xinran Hao (PhD Student)
- Heleni Singer (PhD Student)
- Manini Jain (PhD Student)
- Steffi Spitznagel (PhD Student)
- Toby Curless (Student)
- Doga Yorulmaz (Student)
- Zaynah Shahzad (Student)
- Shahryar Alavi (Research Assistant)
Collaborators






Lab funders
Thank you to all those who support the Hardy Lab!
