Biography
Marc-David Ruepp is a Professor of RNA Biology and Molecular Neurodegeneration at the UK Dementia Research Institute (UK DRI) at King’s College London. His laboratory is interested in RNA metabolism in health and disease with a specific focus on neurodegeneration. He received his PhD in 2009 (Graduate School of Cellular and Biomedical Sciences, University of Bern) and started his independent scientific career as Junior Group Leader in 2014 in the Department of Chemistry and Biochemistry at the University of Bern, Switzerland. He habilitated at the Faculty of Science, University of Bern, and received his Venia Docendi in RNA biology in 2018. The same year he joined the UK DRI as Group Leader and King’s College London as Senior Lecturer in Neuroscience. In 2022 he was promoted to Reader and in 2025 to Professor of RNA Biology and Molecular Neurodegeneration.
Ruepp Lab
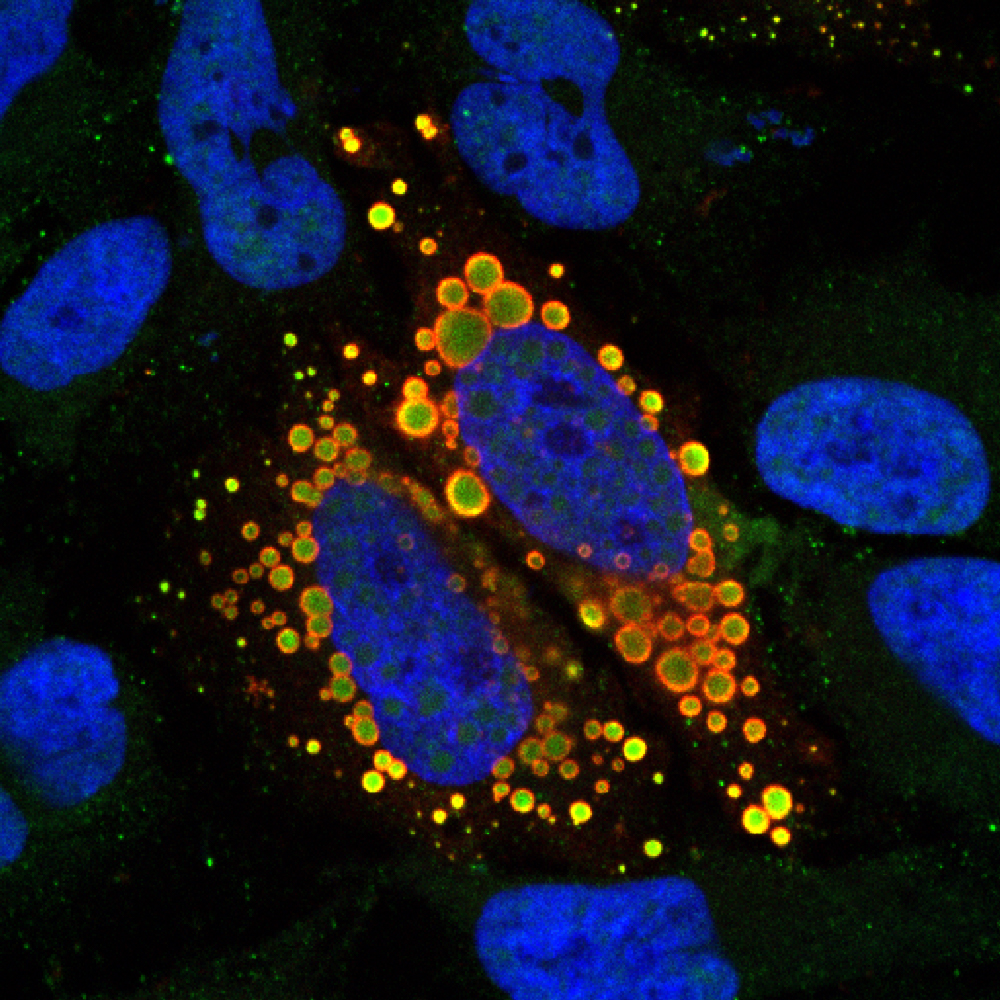
Explore the work of the Ruepp Lab, exploring RNA metabolism in health and disease with a specific focus on neurodegeneration