Key details
Deciphering RNA to rescue motor neurons
In recent decades, scientists discovered that many cases of motor neuron disease/ALS and frontotemporal dementia are caused by problems with proteins that control RNA - an essential molecule for cell function. In particular, proteins called TDP-43 and FUS can malfunction, leading to motor neuron death.
The Ruepp lab investigates exactly how these faulty proteins, especially FUS, cause disease. Their unique approach combines different research models to understand which specific features of FUS are toxic to motor neurons and what cellular processes go wrong as a result.
The team aim to find new treatment targets that could help everyone living with MND/ALS. While some cases are inherited due to gene mutations, 90% occur without a clear genetic cause. Yet all types of ALS result in motor neuron death, suggesting common disease mechanisms. The lab uses stem cell models with different ALS-causing mutations to identify these shared pathways.
Most excitingly, the Ruepp Lab is already testing potential new therapies based on their discoveries. By understanding the fundamental biology of how RNA-binding proteins contribute to motor neuron death, they aim to develop treatments that could benefit both people affected by MND/ALS and FTD .
Latest news

Marc-David Ruepp
Marc-David Ruepp is a Group Leader at the UK DRI at King's. Find out more about his career and expertise on his profile page.

Research summary
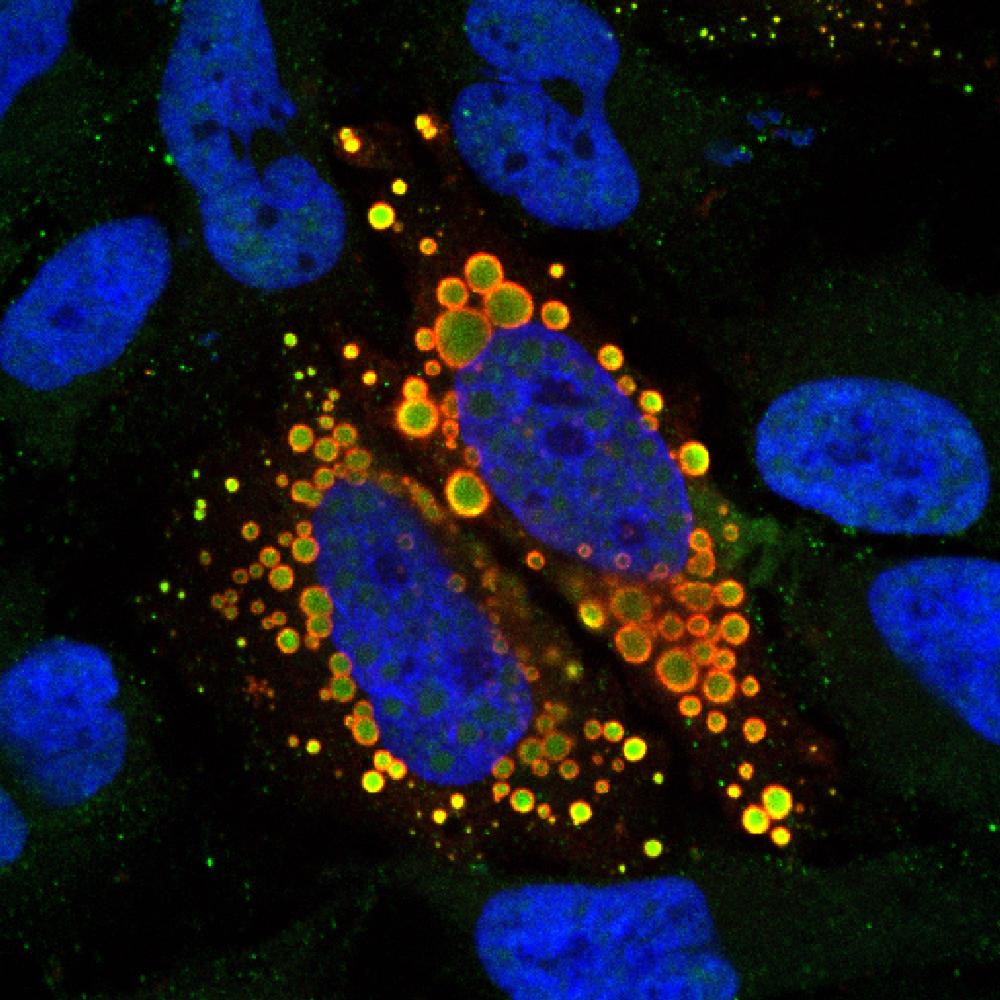
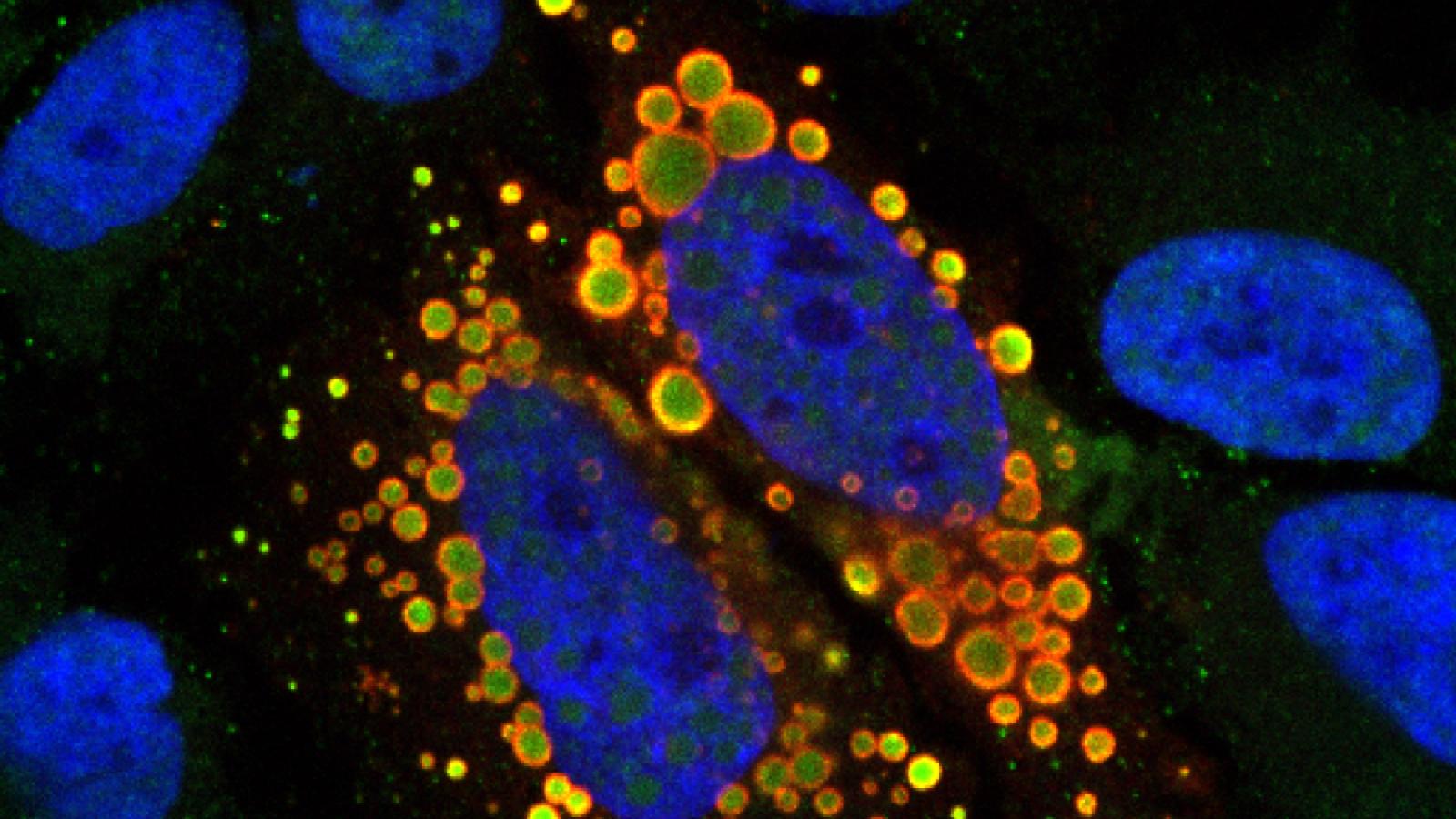
Cytoplasmic mutant TAF15 (red) engulfs U12 snRNAs (green) in the cytoplasm of HeLa cells.
Credit: Tatjana Zoller, Ruepp Lab
Targeting altered RNA metabolism in neurodegeneration
During the last three decades, a variety of neurodegenerative diseases were discovered to be caused by mutations in RNA-binding proteins and altered RNA metabolism appeared on the stage of potential pathomechanisms in neurodegeneration. Amyotrophic Lateral Sclerosis (ALS) and Frontotemporal dementia (FTD) are extreme ends of the ALS-FTD disease spectrum and represent such progressive, fatal neurodegenerative diseases in which altered RNA metabolism is strongly implicated.
Many cases of ALS/FTD are caused by mutations or mislocalisation of RNA-binding proteins such as TDP-43, FUS, as well as by hexanucleotide repeat expansions in the C9ORF72 gene, resulting in the expression of toxic hexanucleotide repeat RNA. The importance of RNA metabolism for neuronal homeostasis is further implicated by the fact that cytoplasmic inclusions of the RNA-binding proteins TDP-43 and FUS are present in approximately 98% of ALS and 54% of FTD cases. While TDP-43 mislocalisation is caused in rare cases by mutations, most cases of TDP-43 pathology are idiopathic. Conversely, FUS-linked ALS arises from mutations in the FUS gene and causes early onset ALS. These mutations typically disrupt the nuclear localisation signal (NLS), leading to cytoplasmic mislocalisation followed by the formation of FUS aggregates in neurons and glial cells. While nuclear loss-of-function has been clearly linked to ALS with TDP-43 pathology, mutant FUS causes motor neuron degeneration via a cytoplasmic toxic gain-of-function. However, the molecular determinants of this toxicity are unknown.
Main objectives and research goals
- Elucidating molecular determinants of FUS toxicity in ALS: We investigate these determinants using in vitro and in vivo disease models combined with rationally designed point mutations to dissect which features of FUS contribute to cytoplasmic toxicity and which cellular pathways are dysfunctional as a direct consequence thereof.
- Identifying translational targets for ALS independent of disease aetiology: Although ALS is heterogeneous with several genes being implicated in disease aetiology, 90% of all ALS cases are sporadic without clear genetic linkage. Nonetheless, all mutations and sporadic cases converge in selective motor neuron death, implicating a shared pathomechanism. Therefore, we are generating and using isogenic de-novo ALS iPSC models across multiple ALS genes to identify converging pathways and novel targets.
- Testing novel targets and approaches for translation: Based on our insights gained from the above work packages, we work towards elucidating the therapeutic and translational potential of new targets and approaches and have started our first pre-clinical studies for ALS/FTD.
Specific techniques
- Splicing modulation using modified snRNAs
- Liquid-liquid phase separation assays
- Microscale Thermophoresis
- qRT-PCR
- Immunoprecipitation
- Affinity purification of proteins/protein complexes
Key publications
Vacancies
Lab members
- Dr Niamh O’Brien (Postdoctoral Researcher - jointly with Dr Sarah Mizielinska)
- Dr Deepak Khuperkar (Postdoctoral Researcher)
- Dr Emma Dyke (Postdoctoral Researcher - jointly with Dr Caroline Vance)
- Richard Taylor (Postdoctoral Researcher)
- Juan Alcalde Gomez (PhD Student)
- Eleanor Wycherley (Research Assistant - jointly with Dr Sarah Mizielinska)
- Dr Christoph Schweingruber (Research Fellow)
- Dr Alessandro Galgiliardi (Research Fellow - with Pietro Fratta)
- Dr Daniel Solomon (Lady Edith Wolfson MNDA Junior Non-Clinical Research Fellow)
- Yujing Gao (Research Associate)
- Michaela Barioglio (Research Associate - with Pietro Fratta)
- Alexander Hofer (PhD Student - with Prof Oliver Muehlemann)
- Vaishnaivi Manohar (PhD Student - with Dr Jemeen Sreedharan)
- Sara Tacconelli (PhD Student - with Dr Caroline Vance)
- Sofia Konstantinidou (PhD Student - jointly with Dr Sarah Mizielinska)
- Emma Salmela (PhD Student - jointly with Prof Chris Shaw)
- Hamish Crerar (Senior Researcher)
Collaborators
















Lab funders
Thank you to all those who support the Ruepp Lab!