Key details
Untangling the effects of tau
Tau is implicated in many different neurodegenerative diseases. The protein normally helps provide structural support to brain cells, acting like a scaffold. In some diseases, known as 'tauopathies', tau can become misfolded and accumulate into clumps or tangles. This causes cells to become unhealthy and die, leading to symptoms such as cognitive decline, or problems with movement.
The gene that codes for tau is very large and complex, and it can vary greatly among different populations around the world. Tau can also behave differently in different diseases, and can form clumps in many different brain regions and cell types.
The Bowles Lab are using cellular models of tauopathies to try and understand what happens within our cells when there is a mutation in the tau gene. Advancing our understanding of tau in this way would provide an important piece of the puzzle that could lead to effective new treatments targeting diseases involving tau.
Latest news


Dr Kathryn Bowles
Dr Kathryn Bowles is a Group Leader at the UK DRI at Edinburgh. Find out more about her career and expertise on her profile page

Research summary
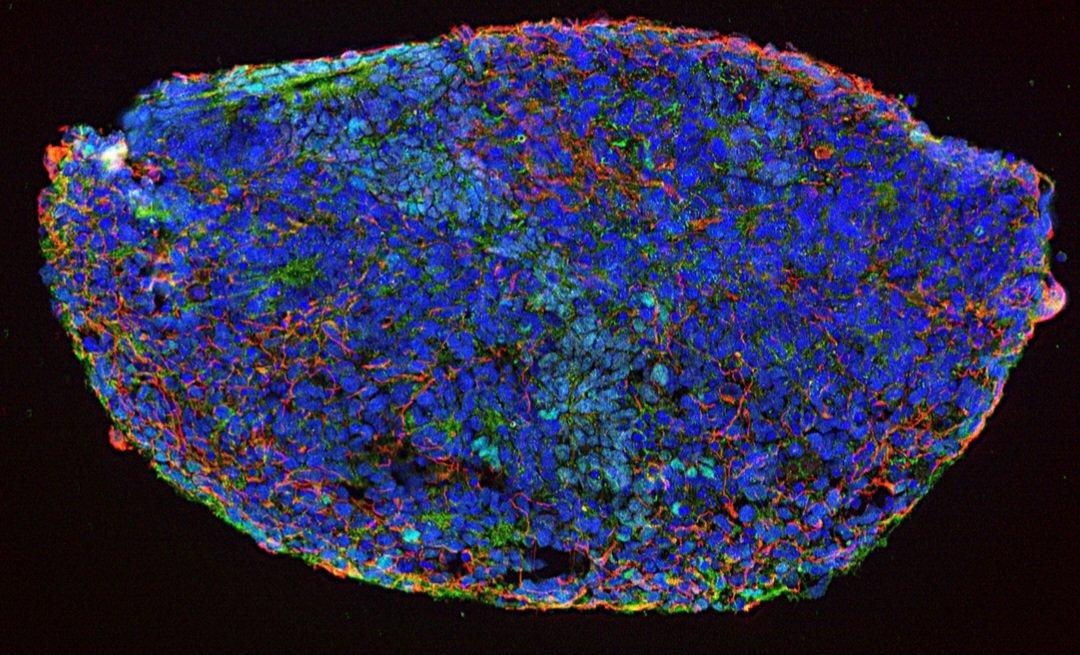
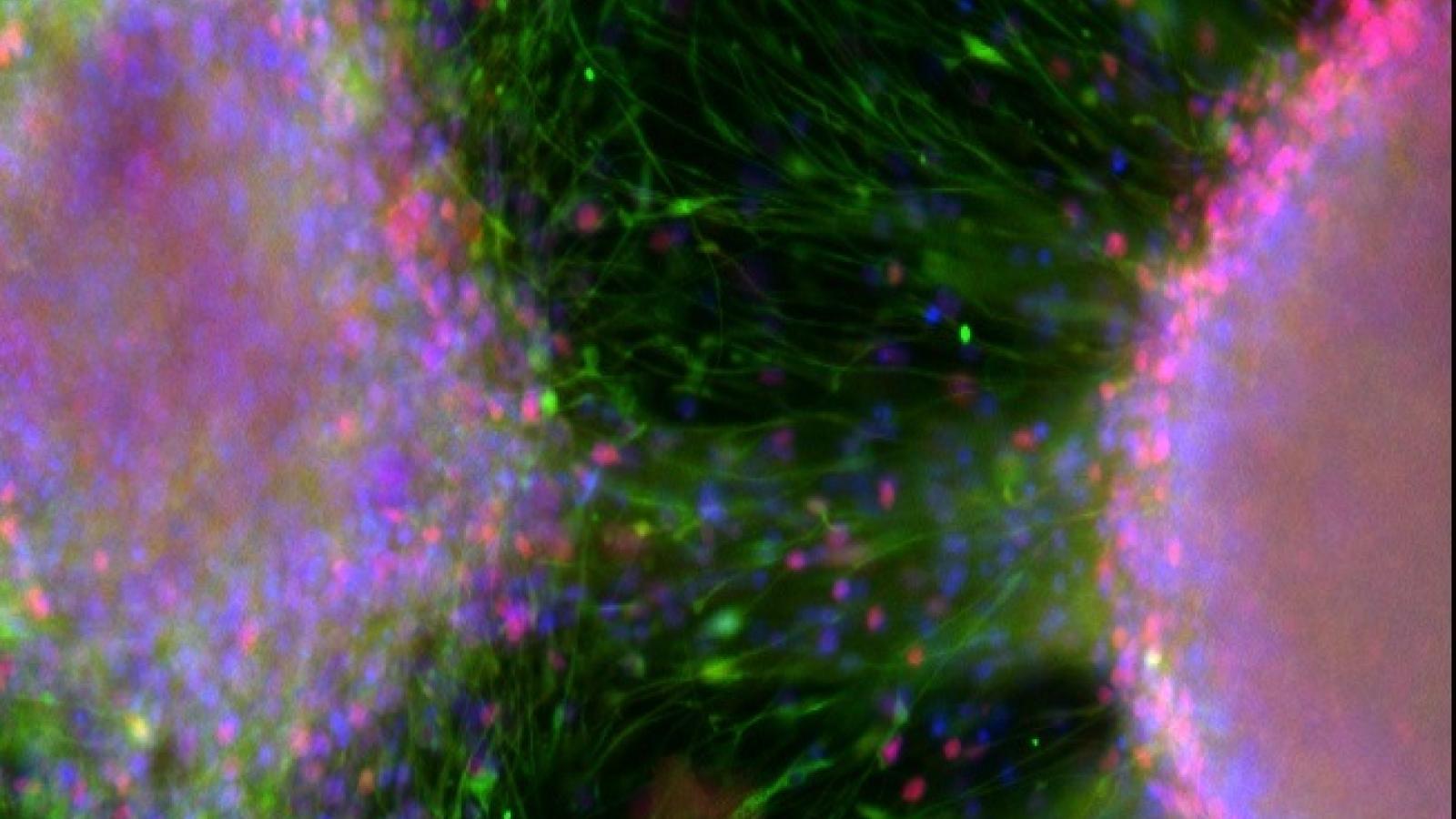
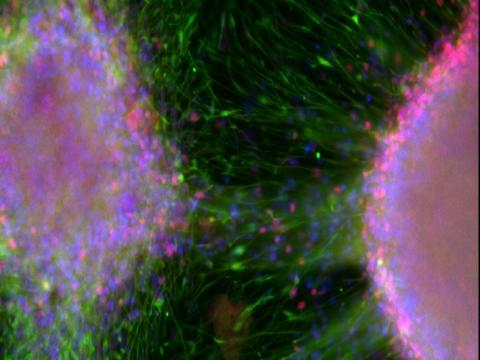
Immunofluorescence image of cryosectioned organoid, with DAPI in blue, b3-tubulin in red and FOXG1 in green. Credit: Filipa Henderson Sousa/Kathryn Newton, Bowles Lab
Genetic mechanisms underlying tauopathies
Frontotemporal dementia (FTD) encompasses a spectrum of disorders affecting behaviour and speech, and accounts up to 20% of dementia cases under age 65. Mutations in the microtubule-associated protein tau (MAPT) gene have been identified in up to 38% of familial FTD cases (FTD-tau). iPSC-based models derived from patients with MAPT mutations show promise for studying early molecular mechanisms underlying tauopathy and recapitulate many disease-associated phenotypes observed in human brain. Dr Bowles's research employs iPSC modelling to investigate the early molecular mechanisms underlying FTD-tau.
In previous work, the Bowles Lab characterized a 3D cerebral organoid model of FTD-tau, with the goal of identifying temporal changes preceding and leading to neurodegeneration. They found that this model exhibited progressive accumulation of total and phosphorylated tau, and later specific loss of glutamatergic neurons. The team are now investigating the role of glutamate signalling in synaptic function and neuronal survival in an organoid model of FTD.
The 4R isoform of tau is important in FTD and progressive supranuclear palsy (PSP) pathology, but it is challenging to model using iPSC systems, as neurons derived from these cells express very low levels of 4R tau. Kathryn and her team have developed a panel of iPSC lines, derived from donors with specific MAPT mutations, that express significantly more 4R tau than controls. The researchers are using these iPSC lines to model the early changes occurring in PSP/FTD brain due to the increased expression of 4R Tau, in order to better understand the functional impact of this isoform in multiple cell-types, with a goal to inform future therapeutic development. This includes the assessment of both astrocytes and microglia, and how they affect neuronal health in the context of tauopathy.
Due to the genetic association with the 17q21.31 (MAPT) locus, Kathryn examined genetic data from a cohort of patients with Parkinson’s, and identified a novel gene, LRRC37A2 as reducing risk. Specifically, higher LRRC37A2 expression in astrocytes due to increased copy number was associated with protection against Parkinson’s. The lab are now using iPSC models to examine the function of LRRC37A2 in depth in order to uncover how it may be beneficial for people with Parkinson’s.
Main objectives:
- Use iPSC modelling to investigate the early molecular mechanisms underlying tauopathy.
- Investigate the role of glutamate signalling in synaptic function and neuronal survival in a model of FTD.
- Understand the cell autonomous and non-cell autonomous effects of tau expression in glia
- Determine the function of LRRC37A2 and identify novel mechanisms of protection for Parkinson’s.
Key publications
Vacancies
Lab members
- Kathryn Newton (Senior Research Technician)
- Filipa Henderson-Sousa (Postdoctoral Researcher)
- Dianne Lopez (Postdoctoral Researcher)
- Sam Heron (Postdoctoral Researcher)
- Dafni Kampitsi (PhD Student)
- Paula Beltran-Lobo (Postdoctoral Researcher - jointly with Blanca Díaz Castro)
- Lois Keavey (Research Assistant)
- Trinity Rungasamy (Research Assistant)





Collaborators










Lab funders
Thank you to all those who support the Bowles Lab!