Identifying the causes of motor neuron disease
Amyotrophic lateral sclerosis (ALS, also known as motor neurone disease (MND)) and frontotemporal dementia (FTD) are neurodegenerative diseases, which emerging evidence shows may have significant overlap in biological cause. Although most people who develop ALS have no family history of the disease, around 5-10% of cases are inherited – with up to 40% due to a fault in a gene known as C9orf72. This faulty gene is also the most common genetic cause of FTD.
In healthy individuals, the DNA sequence of C9orf72 contains a short recurring repeat of 6-letters. But in people with a faulty copy, this is hugely expanded and leads to accumulation of abnormal proteins. The Chandran Lab are studying the biological consequences of these so-called ‘repeat expansions’ in the C9orf72 gene on different types of brain cell, particularly immune cells that support neurons.
The Chandran Lab aim to tease out the root causes of ALS and/or FTD and identify ways to help protect neurons from damage. For example, they will explore if an accumulation of the mutant gene sequence within cells is toxic, or alternatively, if problems arise due to low levels of the functional C9orf72 protein. The team hope this work will lead to effective new treatments in the future.
Latest news


Prof Siddharthan Chandran
Prof Siddharthan Chandran is Director & CEO of the UK Dementia Research Institute, and Group Leader at the Edinburgh and UCL Centres. Find out more about his career and expertise on his profile page.

Research summary
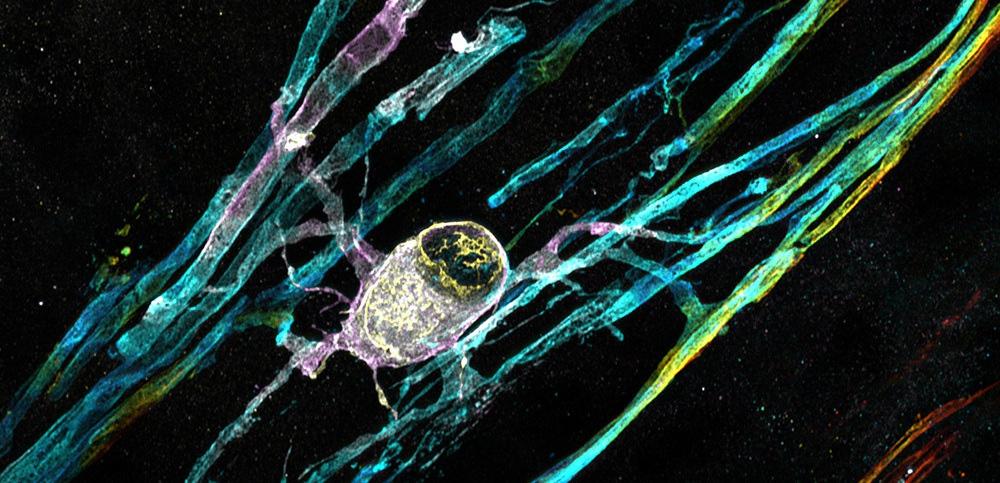
Human stem cell-derived myelinating oligodendrocyte can be seen with many myelinating processes wrapped around unstained neurons Credit: Dr Owen Gwydion James, Prof Siddharthan Chandran’s lab, UK DRI at Edinburgh
Dissecting macroglial-neuronal crosstalk in C9ORF72 FTD/ALS
The overall objective of this UK DRI programme, led by Prof Siddharthan Chandran, is to better understand glial-neuronal cross talk and how this is altered in neurodegenerative disorders, using patient-derived mutant C9orf72 iPS cell lines as a prototypic example, and determining whether manipulating axo-glial interaction is neuroprotective.
The C9orf72 mutation (mC9) is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). How repeat expansions in the C9orf72 gene (C9) cause these conditions is unknown. Three broad mechanisms, not mutually incompatible, are offered: loss of function of C9 protein, gain of function toxicity via C9 repeat RNA foci or dipeptide repeats (DPRs) generated by non-ATG translation of repeats. Numerous cellular processes, many also found in both sporadic and familial ALS/FTD, have been identified. Any unifying mechanism will need to also account for the finding of mis-accumulation of insoluble TDP-43, an RNA-binding protein, in >97% of ALS cases.
Until recently, a neurocentric approach has dominated the study of neurodegenerative disease, including ALS/FTD, and yet multiple lines of evidence show glia pathology including accumulation of TDP-43 and C9 RNA foci in ALS. This programme’s focus is on defining the molecular, cellular and functional cell and non-cell autonomous consequences of mC9 mutation on glia.
The team’s approach combines in vitro and in vivo (chimeric transplants) experiments using 3 independent mutant patient iPS cell lines plus isogenic gene-corrected pair to derive neurons (cortical and MN), astrocyte, oligodendrocyte and microglia. Current studies remain focused on first confirming and defining key phenotypes, with follow up mechanistic experiments guided by unbiased approaches such as transcriptomics and proteomics.
Main aims and research goals:
- To determine cell autonomous consequences of C9orf72 mutation on cortical and motor neurons. The team are systematically mapping the cellular and biochemical pathology of cortical and motor neurons – to identify unique and overlapping transcriptomal neuronal signatures and discover regulatory processes that potentially interact with glia signalling pathways.
- To determine the molecular, cellular and functional cell and non-cell autonomous consequences of C9orf72 mutation on cortical and spinal astrocytes. The team are defining the kinetics and identity of astrocyte-mediated factors that result in non-cell autonomous neuronal pathophysiology – they are examining the in vivo impact of age and mutant astrocytes on neuronal health using chimeric transplant experiments.
- To determine the molecular, cellular and functional cell and non-cell autonomous consequences of C9orf72 mutation on cortical and spinal oligodendrocyte precursor cells (OPCs) and oligodendrocytes (OLGs). The team are defining the kinetics and identity of OLG mediated factors that result in non-cell autonomous neuronal pathophysiology – using transplant experiments to evaluate myelination efficiency and the in vivo impact of age and mutant OLGs on neuronal health.
Key publications
Vacancies
Lab members
- Dr Karen Burr (Project Officer)
- Dr David Hampton (Academic)
- Dr Roderick Carter (Postdoctoral Researcher)
- Dr Marcus Keatinge (Postdoctoral Researcher)
- Dr Vidya Ramesh (Postdoctoral Researcher)
- Dr Arpan Mehta (Postdoctoral Researcher)
- Dr Nhan Pham (Postdoctoral Researcher)
- Dr Paul Baxter (Postdoctoral Researcher)
- Dr Laura Planas Serra (Postdoctoral Researcher)
- Dr Sophie Hawkins (Postdoctoral Researcher jointly with Bhuvaneish Selvaraj)
- Esra Ozkan (Postdoctoral Researcher jointly with Bhuvaneish Selvaraj)
- Dr Mungo Harvey (Postdoctoral Researcher jointly with Bhuvaneish Selvaraj)
- Dr Daria Iakovleva (Postdoctoral Researcher jointly with Bhuvaneish Selvaraj)
- Dr Reiss Pal (Posdoctoral Researcher jointly with Bhuvaneish Selvaraj)
- Dr Johnny Tam (Clinical Research Fellow)
- Rebecca Hughes (Data Scientist)
- Dr Raja Nirujogi (Visiting Scientist)
- Dr Gemma Sullivan (Research Fellow)
- Denise O'Keefe (Senior Technician)
- James Cooper (Senior Technician)
- Karen Gladstone (Senior Technician)
- Jyoti Nanda (Senior Technician)
- David Story (Senior Technician - Lab manager)
- Orla Maryland (Technician)
- Emily Yates (Technician)
- Cameron Henn (Technician)
- Sandra Constantinou Juhasz (Technician)
- Dr Maria Stavrou (PhD Student)
- Abby O'Sullivan (PhD Student)
- Jade Lucas (PhD Student - jointly with Bhuvaneish Selvaraj)
- Hatice Bozkurt (PhD Student - jointly with David Hunt)
- Steph Panol (PA)
Collaborators











Lab funders
Thank you to all those who support the Chandran Lab!